Dr. Shramin Key Research Areas
Advanced Analytical Techniques
Dr. Mst Sharmin Aktar specializes in developing, validating, and troubleshooting analytical methods for characterizing drug substances and formulations. Her expertise includes HPLC, UV-Vis spectrophotometry, and FTIR, ensuring compliance with cGMP, FDA, and ICH guidelines. She has a strong background in method transfer, assay qualification, and process optimization to enhance laboratory efficiency and data integrity.
Protein Purification & Structural Biology
Dr. Aktar has successfully developed and optimized protein purification protocols using chromatography-based techniques. Her research in structural biology led to the analysis and publication of five novel protein X-ray crystal structures in the RCSB Protein Data Bank. These findings contribute to the understanding of enzyme mechanisms, protein-ligand interactions, and biocatalysis, advancing drug discovery and therapeutic protein development.
Formulation & Drug Development
With extensive experience in NDA/ANDA solid dosage formulation development, Dr. Aktar has played a pivotal role in designing and optimizing stable, bioavailable, and scalable pharmaceutical formulations. She has successfully led technology transfer, regulatory compliance efforts, and process improvements, ensuring strict adherence to FDA, MHRA, and ICH guidelines. Her contributions support drug development, quality control, and regulatory submissions in GMP-regulated environments.
Explore Dr. Sharmin Latest Publications
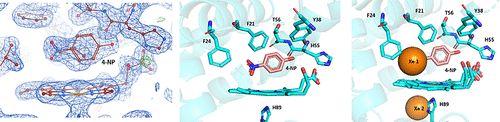
Decoding Enzyme-Substrate Interactions for Biochemical Advancements
This study explores the structural differences in substrate binding between Dehaloperoxidase A (DHP A) and Dehaloperoxidase B (DHP B) using X-ray crystallography and biophysical analysis. The findings reveal key variations in active site architecture that influence substrate recognition and catalytic efficiency, providing deeper insights into enzyme function, ligand specificity, and potential biotechnological applications.
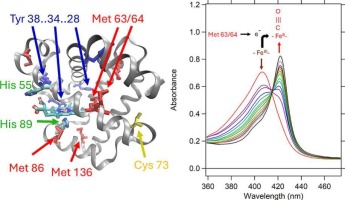
The Hidden Role of Electron Transfer in Biological Reactions
This study examines the mechanisms of proton-coupled electron transfer (PCET) in Dehaloperoxidase (DHP), a heme-containing enzyme. Using spectroscopic and kinetic analysis, the research reveals how electron and proton movements are coordinated to regulate enzymatic activity. The findings enhance our understanding of heme redox chemistry, biological electron transfer, and potential biocatalytic applications.
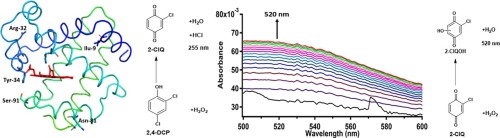
How Enzymes and Hydrogen Peroxide Break Down Environmental Pollutants
This study investigates the oxidative breakdown of 2,4-dihalophenols through both enzyme-catalyzed (dehaloperoxidase) and spontaneous hydrogen peroxide-mediated reactions. Using kinetic and spectroscopic analysis, the research compares the efficiency and pathways of enzymatic vs. non-enzymatic degradation. The findings provide valuable insights into environmental bioremediation, enzymatic oxidation mechanisms, and potential biotechnological applications for pollutant degradation.
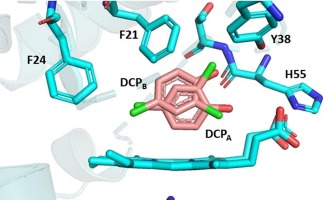
Investigating How Enzyme Variants Process Toxic Chemicals Differently
This study explores the differences in how Dehaloperoxidase A (DHP A) and Dehaloperoxidase B (DHP B) bind and activate 2,4-dichlorophenol, a common environmental pollutant. Using spectroscopic and kinetic analyses, the research reveals distinct enzyme-substrate interactions that influence reactivity, binding affinity, and oxidation efficiency. These findings enhance our understanding of enzyme selectivity in pollutant degradation and potential applications in environmental bioremediation.
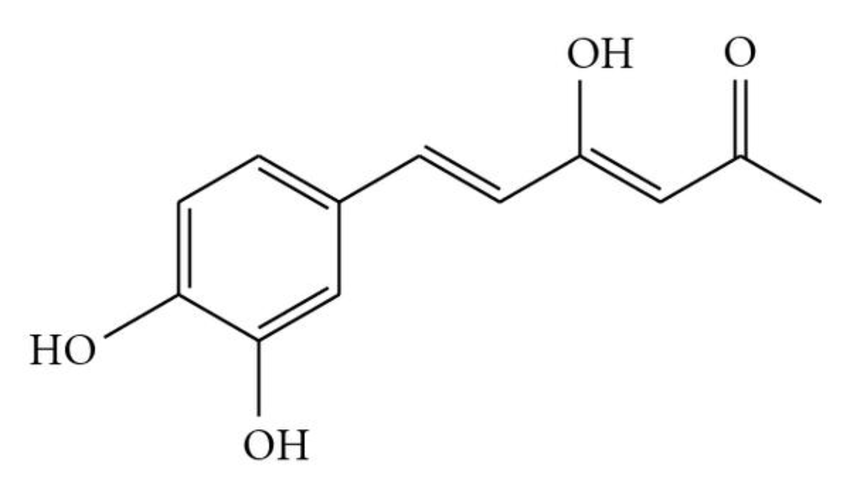
Can Hispolon Be a Safe Drug? A Computational Investigation
This study utilizes computational (in silico) methods to analyze the drug-like properties of Hispolon, a naturally occurring compound with potential therapeutic applications. Using molecular modeling and predictive algorithms, the research evaluates physicochemical parameters, pharmacokinetics (ADMET properties), and toxicity risks. The findings provide valuable insights into Hispolon’s drug potential, absorption, metabolism, and safety, supporting its future applications in drug discovery and development.
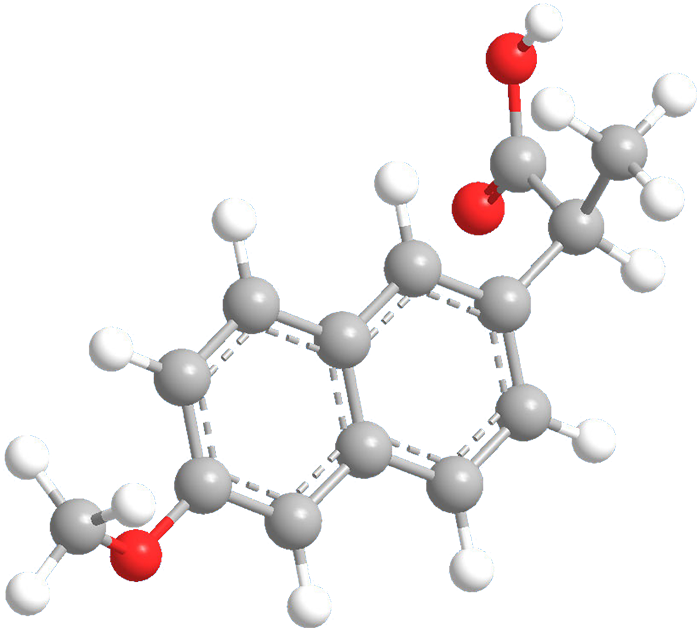
Predicting Naproxen’s Pharmacological Behavior Through Molecular Docking
This study employs computational chemistry techniques to analyze the electronic structure and binding interactions of Naproxen, a widely used nonsteroidal anti-inflammatory drug (NSAID). Using quantum mechanical calculations and molecular docking, the research investigates geometry optimization, polarizability, hyperpolarizability, and receptor binding affinity. The findings provide valuable insights into Naproxen’s structural properties, potential bioactivity, and interaction mechanisms, contributing to drug formulation and optimization.
Dr. Sharmin Protein Structures
Dr. Mst Sharmin Aktar has made significant contributions to structural biology through her research in X-ray crystallography and protein characterization. She has successfully solved and analyzed five novel protein crystal structures, which have been accepted into the RCSB Protein Data Bank.
Her work provides critical insights into enzyme mechanisms, substrate interactions, and catalytic activity, advancing the understanding of protein-ligand binding and structural dynamics. By utilizing high-resolution crystallographic techniques, she has contributed to drug discovery, biocatalysis, and enzyme engineering applications.
Dr. Aktar’s research plays a crucial role in bridging fundamental protein science with pharmaceutical innovations, enabling the development of targeted therapeutics and biotechnological advancements.
Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol
Protein Structure
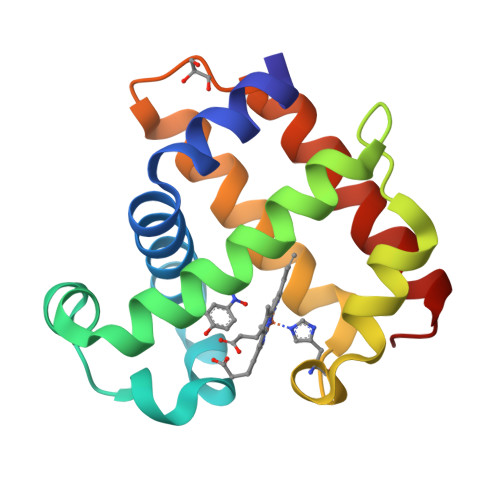
- PDB DOI: https://doi.org/10.2210/pdb8VKC/pdb
- Deposited: 2024-01-08 Released: 2024-07-17
- Funding Organization(s): National Science Foundation (NSF, United States)
Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol
Protein Structure
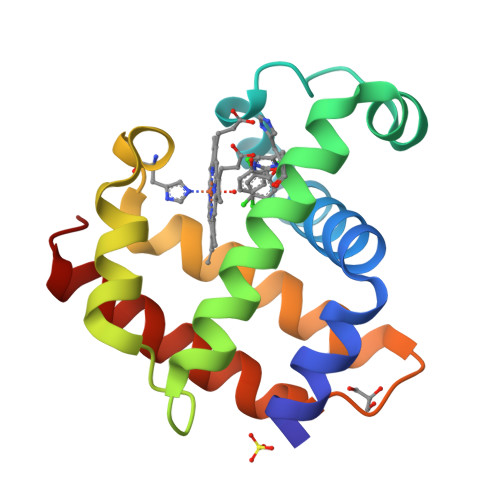
- PDB DOI: https://doi.org/10.2210/pdb8EJN/pdb
- Deposited: 2022-09-17 Released: 2023-08-23
- Funding Organization(s): National Natural Science Foundation of China (NSFC)
Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol
Protein Structure
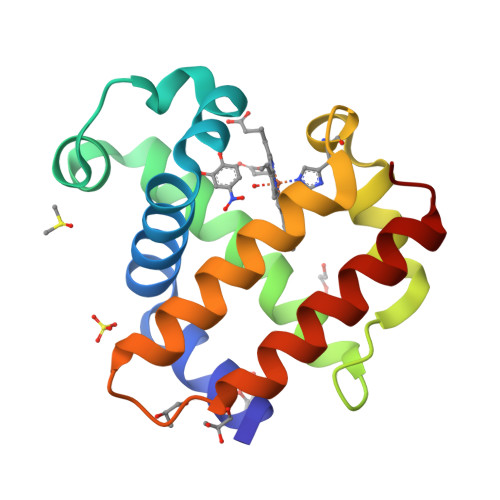
- PDB DOI: https://doi.org/10.2210/pdb8VKD/pdb
- Deposited: 2024-01-08 Released: 2024-07-17
- Funding Organization(s): National Science Foundation (NSF, United States)
Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol
Protein Structure
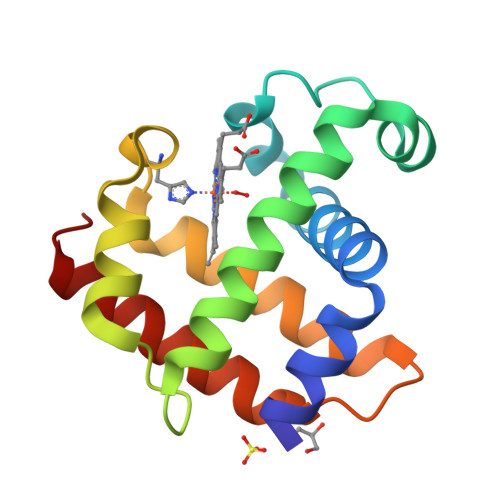
- PDB DOI: https://doi.org/10.2210/pdb8VSK/pdb
- Deposited: 2024-01-24 Released: 2024-07-17
- Funding Organization(s): National Science Foundation (NSF, United States)
Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol
Protein Structure
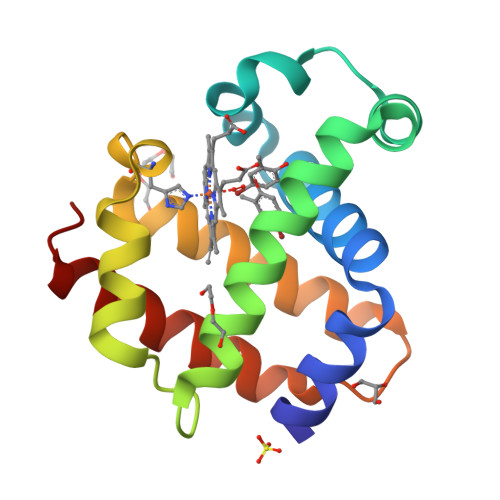
- PDB DOI: https://doi.org/10.2210/pdb8VZR/pdb
- Deposited: 2024-02-12 Released: 2024-07-17
- Funding Organization(s): National Science Foundation (NSF, United States)
